Evolution and Ecology of Invasive Plant Species
Role of Genome Duplication, Climate Change and Natural Enemies in the Range Expansion and Invasibility of Plants
Project Overview
Identifying traits that distinguish invasive from non-invasive species is central to invasion biology and studies indicate that plants with a certain suite of traits can be more successful. Genome characteristics such as ploidy level and genome size (GS) are attributes that influence many traits at different levels of biological organization and are of great interest in ecology and evolution. Evidence suggests that ploidy and GS variation within ploids influence plant traits that in turn, can affect interactions with natural enemies and response to environmental change and ultimately invasiveness. Genomic variation can lead to the spread or decline of a plant species in novel environments. With Laura Meyerson, lead investigator, and a team of international scientists that form PhragNet; see below), our goals were to:
(1) Document variability in GS and ploidy level in Phragmites australis at the local (within site) and regional (spanning 2 continents) scales in field surveys in North America (NA) and Europe (EU).
(2) Using common gardens (Fig. 1) on both continents with plant material collected from mixed cytotype populations, experimentally test functional variability in fitness, phenological, physiological, morphological and natural-enemy defense traits among genetic variants. Also, in the gardens, test predictions that plants with different GS and ploidy exhibit different degrees of plasticity and tradeoffs involving fitness and defensive traits.
(3) Conduct an experiment to assess the effect of ploidy, GS and genotype on intra- and inter-specific competition; a plant trait often linked to invasion success and range expansion.
(4) Investigate the effects of climate-change variables and natural enemies on trait expression for Phragmites variants with different ploidy and GS levels.
(5) Using field surveys, assess how environmental factors influence the distribution and clonal growth of genetic variants and whether the diversity and impact of natural enemies varies among genotypes, ploidies, GS.
(6) Finally, use structural equation models to investigate the combined effects of cytotype, genotype, herbivores, and climate-change variables on plant fitness, range expansion and invasiveness.sing field surveys, assess how environmental factors influence the distribution and clonal growth of genetic variants and whether the diversity and impact of natural enemies varies among genotypes, ploidies, GS.


No prior study has definitively investigated the effects of cytogenetic and genotypic characteristics on functional plant traits at multiple scales. We address the following questions: What is the intraspecific variation in GS and genome copy number within Phragmites and is this structured geographically, by origin and/or invaded region? How do cytological, genetic, ecological and geographical factors interact to determine the population invasiveness at the intraspecific level? What are the direct and indirect effects of these determinants and their relative contribution to determining local and regional patterns of spread and invasion? Three approaches are being used to address these questions: (i) genetic and cytogenetics, (ii) common garden experiments and (iii) field surveys.
Our focal plant, the cosmopolitan species Phragmites australis, is a model system for studying the questions and objectives outlined above (Meyerson et al. 2016). P. australis is adapted to wide climatic and latitudinal ranges (±60°), including extreme environments. It exhibits high genotypic diversity, intra- and interspecific hybridization, globally distributed diverse ploids (4x-12x, 22x) and GS variability within ploidy levels, and a near complete evolutionary history.
Common-Garden Study
Meyerson, L. A., J. T. Cronin, G. P. Bhattarai, H. Brix, C. Lambertini, M. Lučanová, S. Rinehart, J. Suda and P. Pyšek. 2016. Ploidy level and nuclear genome size modify the expression of plant traits and response to herbivory in Phragmites australis. Biological Invasions 18(9): 2531-2549.
We studied the relationship between genome size and ploidy level variation and plant traits for Phragmites australis. Using a common garden in Aarhus, Denmark that contained a global collection of 166 source populations of P. australis, we investigated the influence of monoploid genome size and ploidy level on the expression of P. australis growth, nutrition and herbivore-defense traits and whether monoploid genome size and ploidy level play different roles in plant trait expression.
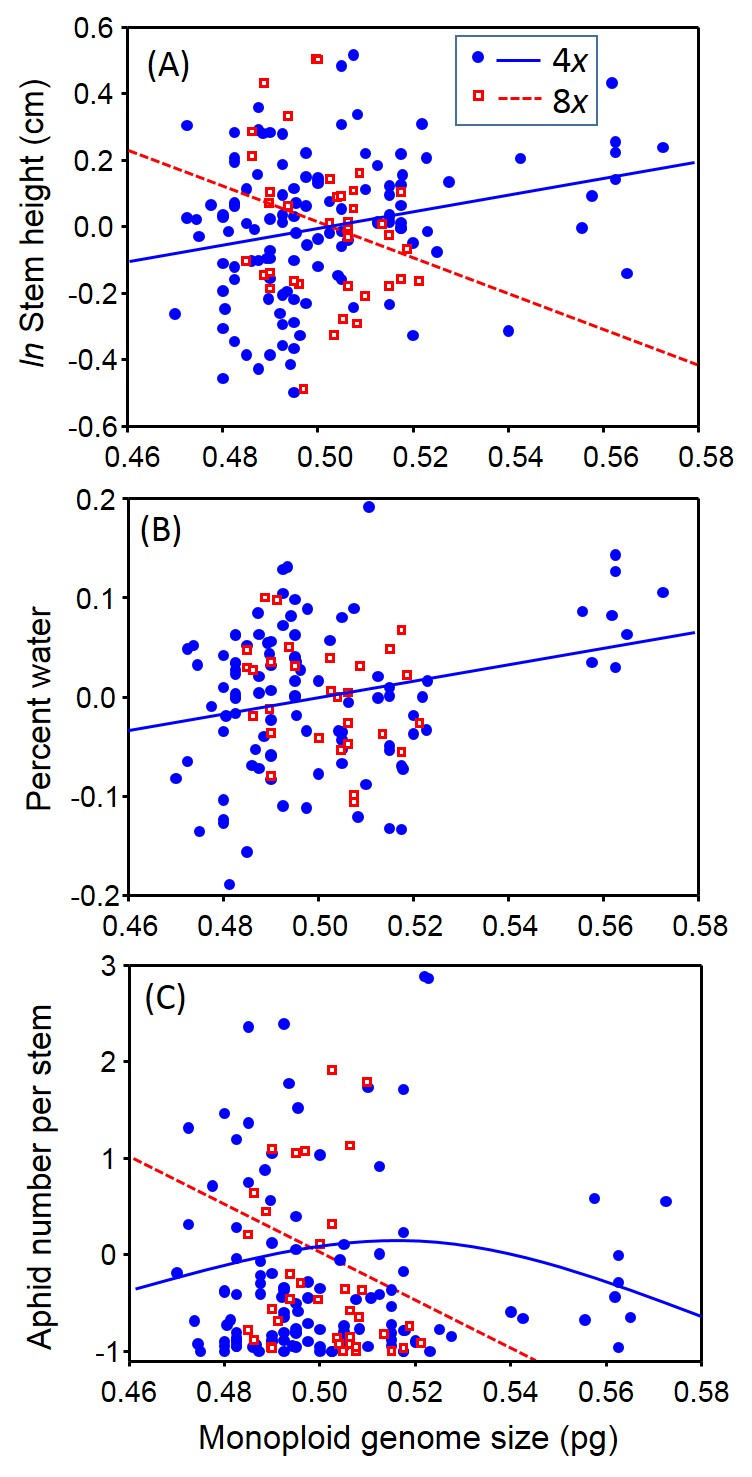
We found that both monoploid genome size and latitude contributed to variation in traits that we studied for P. australis, with latitude being generally better predictor of trait values and that ploidy level and its interaction with monoploid genome size and latitude also contributed to trait variation. We also found that for four traits, tetraploids and octoploids had different relationships with the monoploid genome size. For example, for tetraploids, stem height and leaf water content showed a positive relationship with monoploid genome size, but octoploids had a negative relationship with monoploid genome size for stem height and no relationship for leaf water content (Fig. 2A,B).
As genome size within octoploids increased, the number of aphids colonizing leaves decreased whereas for tetraploids there was a quadratic, though non-significant, relationship (Fig. 2C). Similarly, we found that the relationship between plant traits and latitude depended on ploidy number. Octoploids were sensitive to the latitudinal gradient – stem number increased, water content decreased and palatability to aphids was humped-shaped with increasing latitude (Fig. 3). In contrast, tetraploids showed very little relationship between these same traits and latitude (Fig. 3).
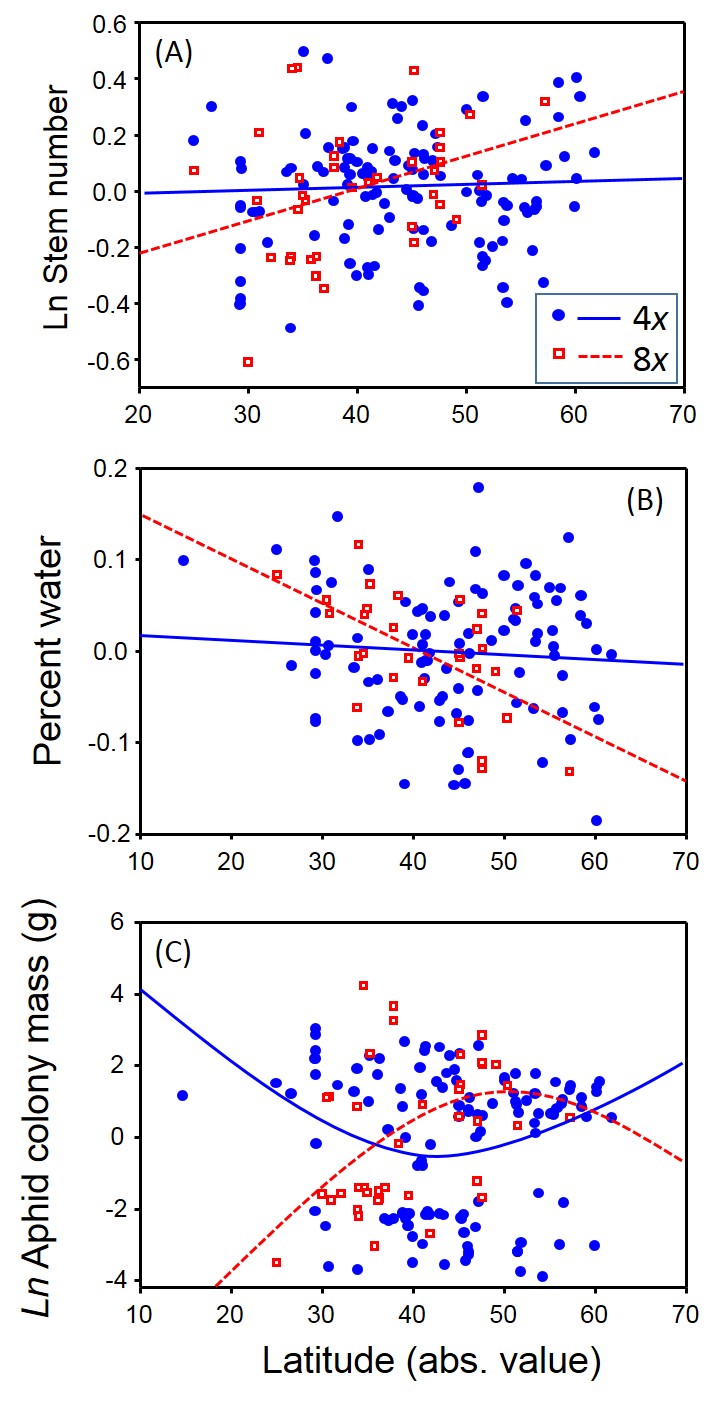
Generally we found that tetraploids were taller, chemically better defended, had a greater number of stems, higher leaf water content, and supported more aphids than octoploids.
This study suggests that there are potential trade-offs among plant traits mediated by monoploid genome size and ploidy with respect to fitness and defense and that the latitude of plant origin is a significant determinant of trait expression even after a decade or more of growing in a common garden setting. Under climate change, some genome size and ploidy variants (both within and among plant species) may more successfully cope with changing external filters (e.g., temperature, salinity, drought), owing to greater phenotypic plasticity and to fitness traits that vary with cytotype. As such, some cytotype and genome size variants may be favored by natural selection, leading to changes in population genome size and/or ploidy structure, particularly at species range limits. Such changes could foster “bottom up” effects and further interact with climate change and distribution of natural enemies that are, and will continue to be, important drivers of range expansions and species invasions.
Subsequently, we conducted another common-garden study in which we experimentally tested the Large Genome Constraint Hypothesis (LGCH)—that plants with large genomes are less tolerant of environmental stress and less plastic under stress gradients than plants with small genomes. We predicted that Phragmites populations with larger genomes would have a lower tolerance and less plasticity to a stress gradient than populations with smaller genomes. We subjected plants from 35 Phragmites populations varying in genome size and lineage to two salinity treatments and two climatic regimes (i.e., greenhouse conditions in Louisiana and Rhode Island).
We found all plant traits, except stomatal conductance, were influenced by environmental stressors and genome size. Increasing salinity was stressful to plants and affected most plant traits. Notably, biomass in the high-salinity treatment was 3.0 and 4.9 times lower for the invasive and native lineages, respectively. Plants in the warmer southern greenhouse had higher biomass, stomate density, stomatal conductance, leaf toughness, and lower aboveground percentage of N and total phenolics than in the northern greenhouse. Moreover, responses to the salinity gradient were generally much stronger in the southern than northern greenhouse. Aboveground biomass increased significantly with genome size for the invasive lineage (43% across genome sizes) but not for the native. For 8 of 20 lineage trait comparisons, greenhouse location 9 genome size interaction was also significant. Interestingly, the slope of the relationship between genome size and trait means was in the opposite direction for some traits between the gardens providing mixed support for LGCH. Finally, for 30% of the comparisons, plasticity was significantly related to genome size—for some plant traits, the relationship was positive, and in others, it was negative. Overall, we found mixed support for LGCH and for the first time found that genome size is associated with plasticity, a trait widely regarded as important to invasion success. For more details, see
Meyerson, L, P. Pyšek, M. Lučanová, S. Wigginton, C.-T. Tran and J. T. Cronin. 2020. Plant genome size influences stress tolerance of invasive and native plants via plasticity. Ecosphere 11: Article e03145.
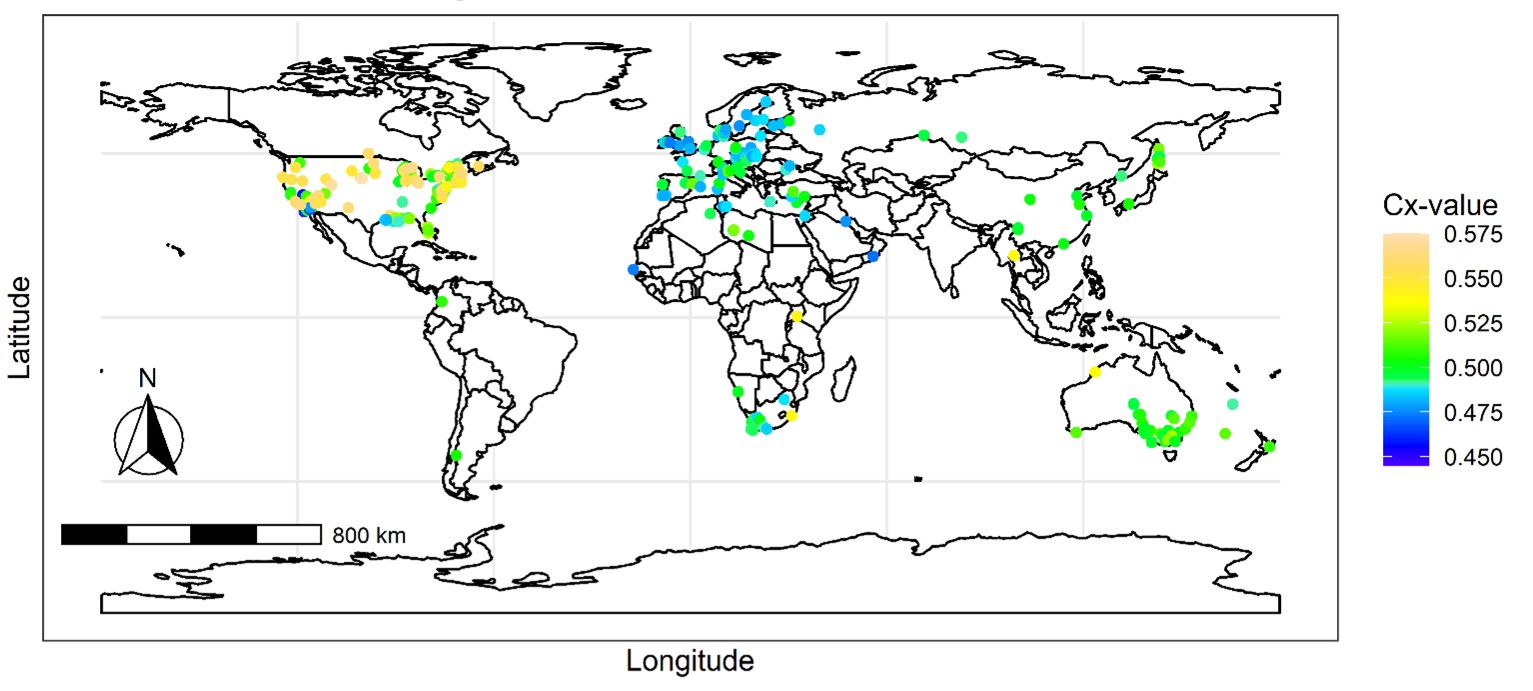
Finally, our same collaborative team used Phragmites australis as a model system to conduct a global analysis of climate and plant native and introduced cytotypes to determine whether this relationship influences population distributions, hypothesizing that smaller genomes are more common in regions of greater environmental stress. First, we identified 598 Phragmites australis field-collected native and introduced genome size variants using flow cytometry (Fig. 4).
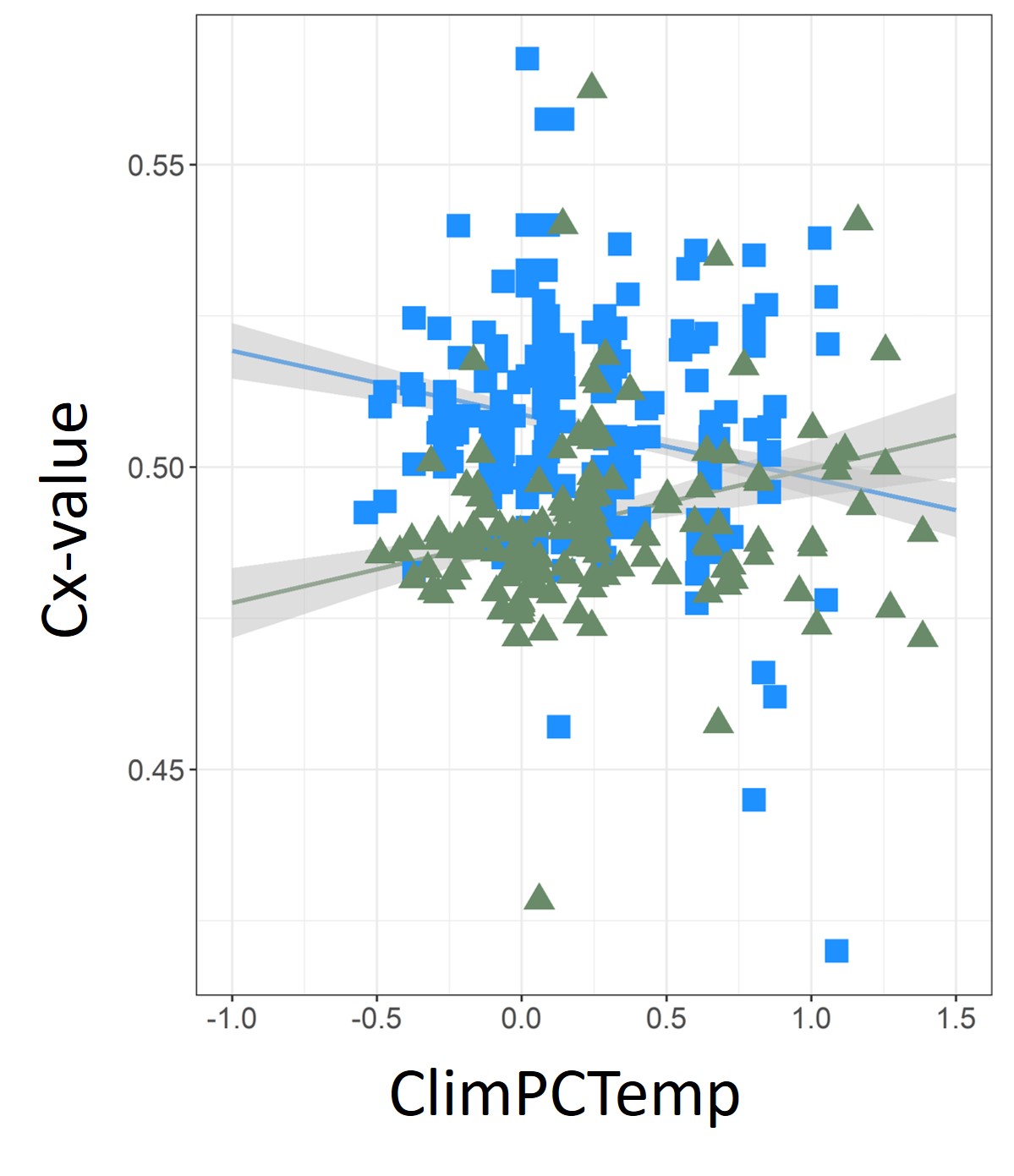
We then evaluated whether temperature and precipitation were associated with P. australis monoploid genome size (Cx-value) distributions using Cx-value and Worldclim data. After accounting for potential spatial autocorrelation among source populations, we found climate significantly influenced Cx-value prevalence on continents. The relationships of Cx-value to temperature and precipitation varied according to whether plants were native or introduced in North America and Europe (see Fig. 5), and Cx-values were strongly influenced by precipitation during the dry season. Smaller plant monoploid genome size was associated with more stressful abiotic conditions; under extreme high temperatures and under drought, plants had smaller Cx-values. This may influence genome dominance, biological invasions, and range expansions and contractions as climate change selects for genome sizes that maximize fitness.
Meyerson, L. A., J. T. Cronin, M. Lučanová, C. Lambertini, J. G. Packer, J. Čuda, J. Wild, J. Pergl and P. Pyšek. 2024. Some like it hot: small genomes are more prevalent under climate extremes. Invasion Biology. 26: 1425-1436.
PhragNet – Global Phragmites Network
In 2013, Meyerson and Cronin, along with Professor Hans Brix (Aarhus Univ., Denmark) and Professor Petr Pyšek (Czech Academy of Sciences, Czech Republic) formed the Global Phragmites Network (PhragNet). This international group of researchers has collaborated on multiple individual research projects in the USA and in Europe since 2010. In 2013, they collectively identified a need to advance the understanding of the effects of global environmental change on plant species distributions, range expansion and plant invasions, and plant-herbivore interactions. All four researcher groups have established active common gardens containing diverse genotypes of Phragmites at their universities, and have begun to exchange plant materials so that a core set of identical clones exist in all four gardens. PhragNet has since expanded to include:
Giuseppe Brundu and Vanessa Lozano, Department of Agriculture, University of Sassari, Italy
Susan Canavan, Center for Invasion Biology, Department of Botany & Zoology, Stellenbosch University, South Africa
Stefano Castiglione and Angela Cicatelli, Department of Chemistry and Biology, University of Salerno, Italy
Melissa McCormick, Smithsonian Environmental Research Center, Maryland, USA
Tom Mozdzer, Department of Biology, Bryn Mawr University, Pennsylvania, USA
Renqing Wang, Shandong University, China
Dennis Whigham, Senior scientist and Deputy Director of the Smithsonian Environmental Research Center, Maryland USA
Jan Čuda and Wen-Yong Guo, Institute of Botany, Czech Academy of Science, Pruhonice, Czech Republic
Franziska Eller and Carla Lambertini, Aarhus University, Aarhus, Denmark
Jasmin G. Packer, School of Biological Sciences, The University of Adelaide, Adelaide, Austalia
PhragNet’s objective is to foster synergistic collaborations in pursuit of the above stated goals. Participation in the network is open to anyone interested in these goals and we expect PhragNet to grow in membership over time.






